Troubleshoothing & FAQ#
Contact#
Public channels#
Private channels#
Please write to Jean-Marc Comby for technical issues and questions about the platform, the API, the database, the utilities, or the MEAs.
For biology and neuroscience-related questions, please write to Dr. Flora Brozzi.
Common issues & FAQ#
Organoids, MEAs and live view#
How can I be sure the organoids are fit for an experiment? What guarantees do you provide on the quality of the organoids? Is neurobiology important ? Do you provide support on the biology side?#
Biocomputing is currently a field of fundamental research, which ideally requires some familiarity with neurobiology in order to understand the current capabilities and limitations of the technology.
If you’d like a detailed introduction, we suggest reading our publication in Frontiers.
We are actively working on making organoids that are fit for the purpose of biocomputing, and we are constantly aiming to improve the quality of the organoids in the following aspects:
Longevity, durability, and stability
Complex, rich activity patterns
Presence of genes known to be relevant in functional learning, neuromodulation, synapses, and other neurobiological processes
Currently, we provide an RNA-seq analysis of relevant genes for the organoids on our website.
Neurobiology is crucial for the success of your experiments, and we are here to help you with any questions you may have on the topic.
Please contact us if you have any doubts about the quality of the organoids, or if you have any questions about the biology side of your experiments, and how it may be relevant to your research.
What are the differences between the MEAs? Does the layout matter?#
We currently use two distinct MEA configurations : 4x8 and 1x32.
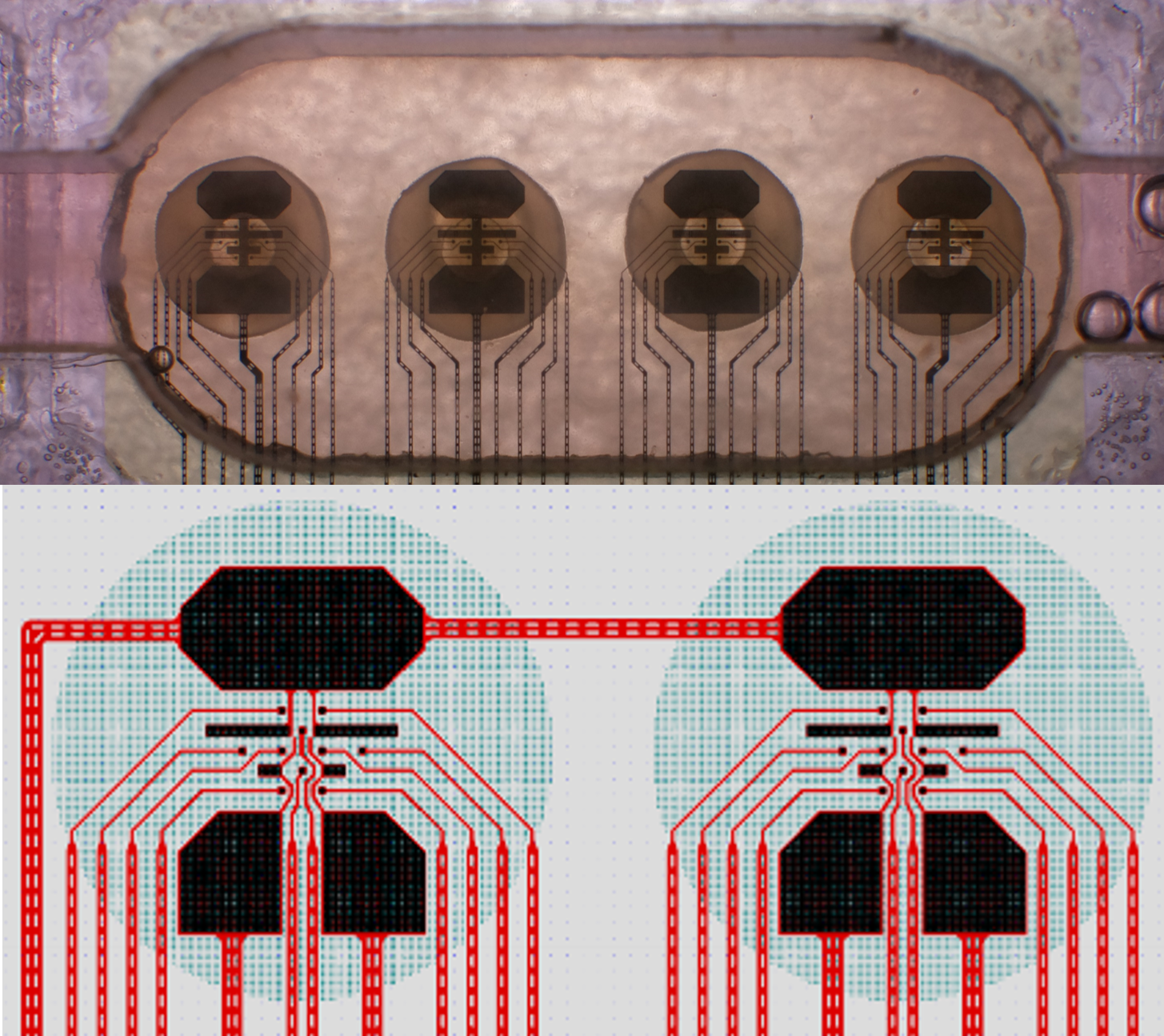
Fig. 7 4x8 MEA configuration, with a close-up of two sites#
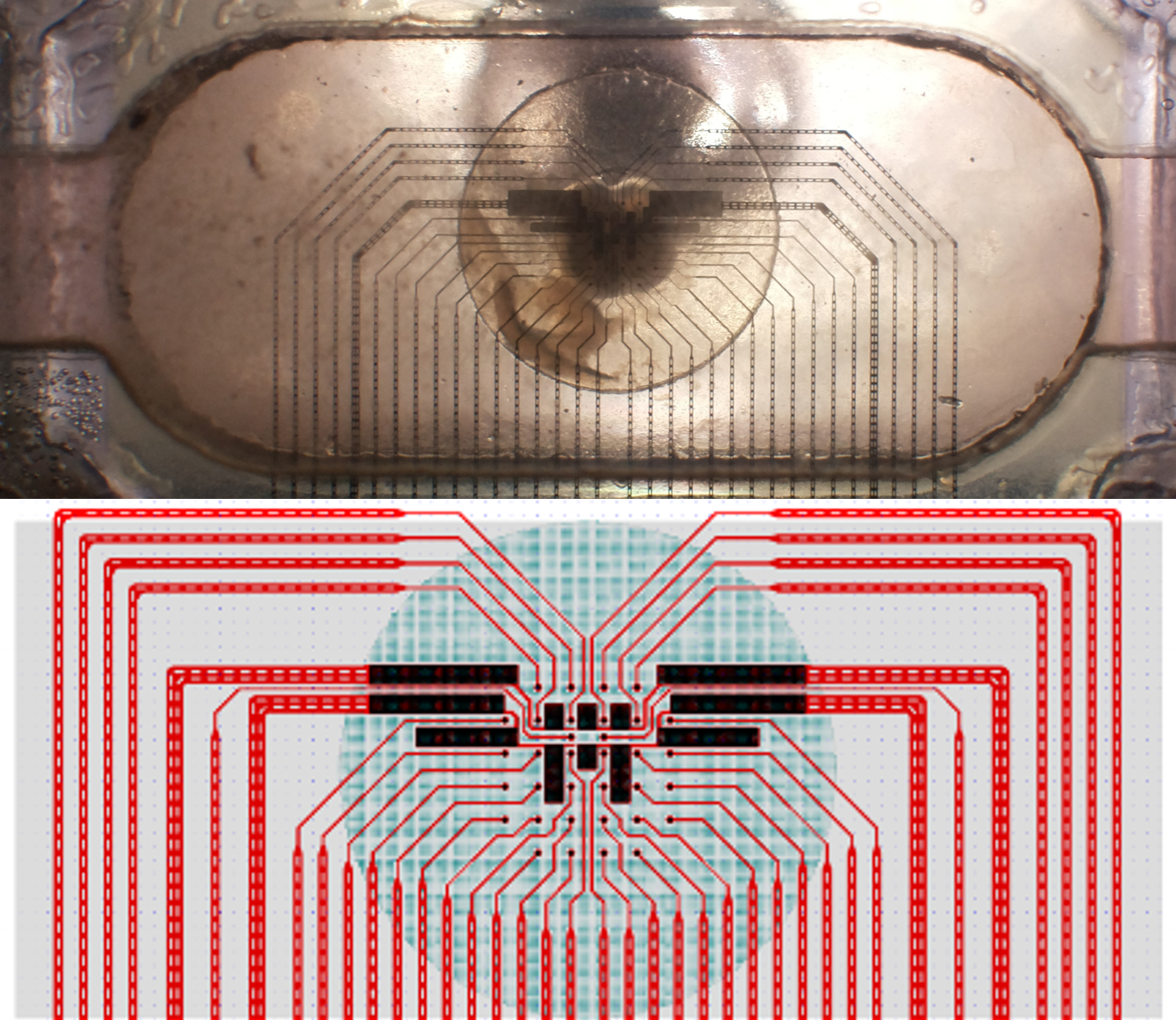
Fig. 8 1x32 MEA configuration#
As there is no cross-talk between sites in the 4x8 configuration, you may parallelize some experiments, which is not possible with the 1x32 configuration (since the sites have cross-talk).
The 1x32 configuration has more sites, which may be useful for some experiments, and may increase the chances of having a particular combination of sites that may be useful for your experiment.
Please contact us if you have further questions about the MEAs and their specifications.
Why is there constant noise on a channel?#
The organoid may not fully cover or be in contact with the electrode, as they are spherical and may not be perfectly centered.
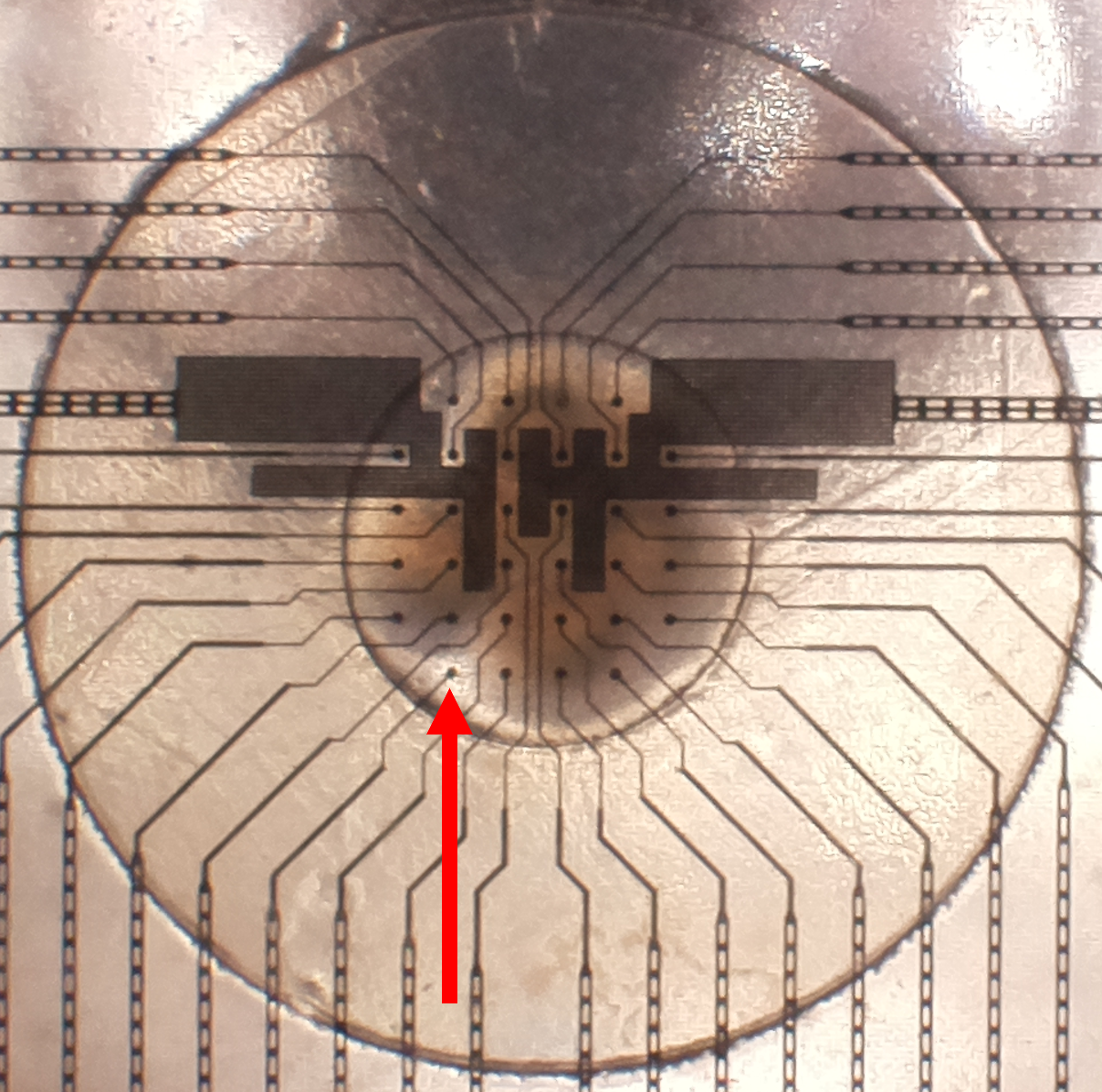
Fig. 9 Partially covered MEA site, highlighted by the red arrow#
There is no signal anymore on the MEA#
This is likely due to the organoids losing activity over time. Please contact us so that we can change the organoids.
Note
If you are using a dedicated access, do not lower the event threshold to get “more activity”, as there may simply not be any activity anymore and you will record noise.
How to differentiate between noise and activity?#
Stimulation artifacts tend to have an amplitude far greater than the organoid’s activity, so checking the amplitude tag of the events may help you determine if there are any artifacts.
Amplitudes below 30 µV are likely to be noise, while amplitudes above 200 to 1000 µV are very likely to be artifacts.
You may check the shape of recorded events in the database, to see if they correspond to the expected shape of spikes.
See the following section for examples of artifacts and spikes shapes.
On a dedicated access, you may inspect the live waveforms to see if they correspond to the expected shape of spikes.
A perfectly regular signal, with a nearly constant amplitude, shape and frequency, is likely to be noise or another extraneous signal, unrelated to the organoid.
Please contact us if you have any doubts; we may also be able to correct or mitigate the issue on our side.
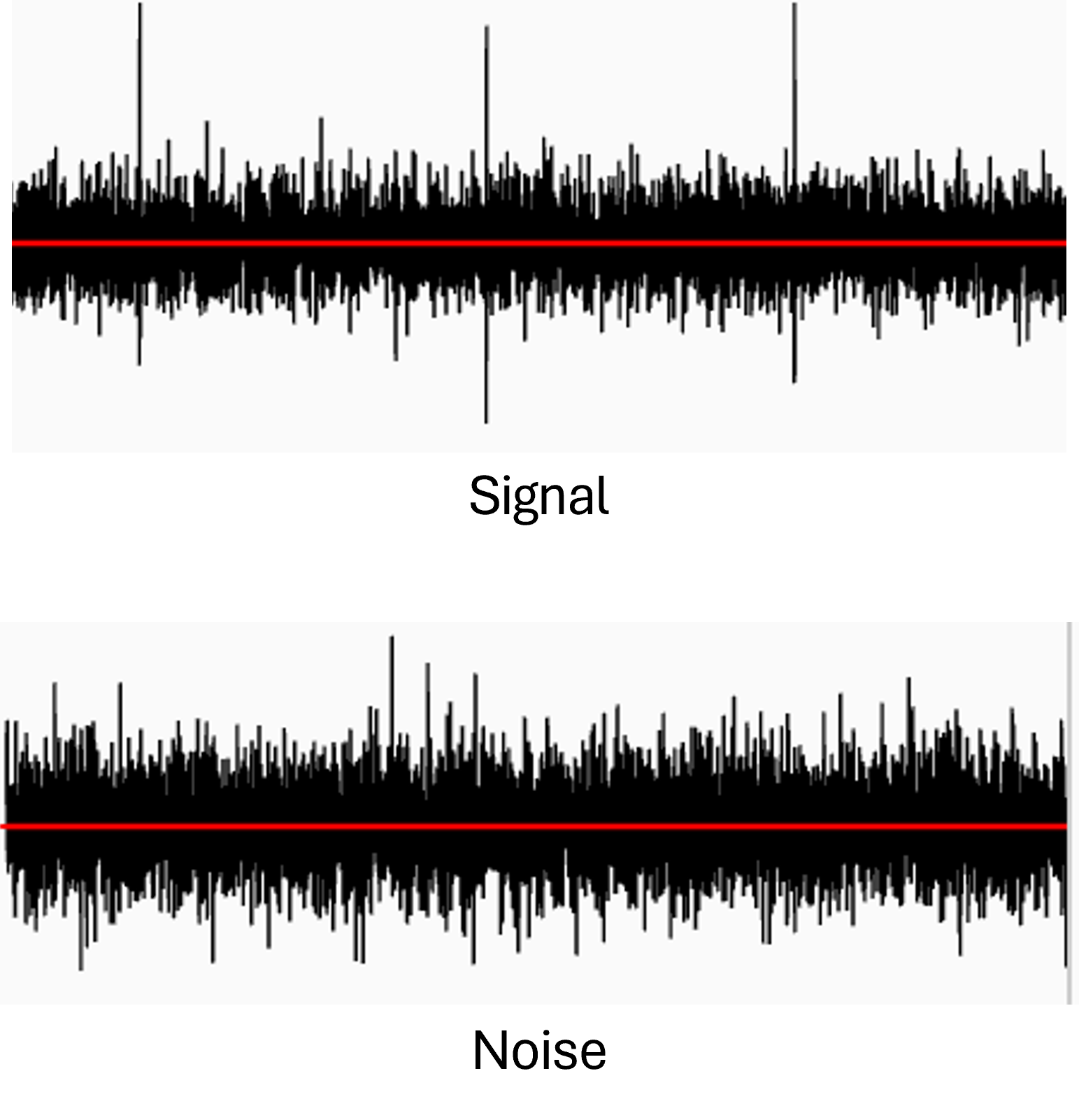
Fig. 10 Example of a signal (top) and noise (bottom) on the same channel from the live view. Scale of +/- 100 µV. Note that the noisy channel shown here is not covered by the organoid.#
The Python API is not responding and/or the live view went into maintenance mode during my booking#
Please contact us.
The spike count on an organoid is too high/low#
Note that “events” are recorded in the database, not spikes.
Since an event is a recording of 3ms whenever the event threshold of \(6\sigma\) is crossed, the number of events is not directly comparable to the number of spikes.
A spiking rate of up to 1500 per minute is still normal (especially in the presence of bursts), but an order of magnitude more would indicate an issue with the event threshold.
For instance, during a burst event, the voltage will nearly always be above the threshold, which will result in an artificial event every 3ms during the burst. However, the many spikes that occur will be merged under several events of 3ms duration. Feel free to ask if you have further doubts regarding the activity of a channel.
Tip
Make sure to reset the variable threshold to True whenever you start or finish an experiment.
See the relevant API section.
Stimulation and parameters#
I am unsure about triggers/parameters, and how I can parallelize my experiments or dynamically change parameters#
Please re-read the system basics page. If anything is still unclear, please contact us.
Some triggers cause stimulation on several sites/MEAs in an unexpected way#
Please make sure to disable all parameters at the end of your experiment, as they are not reset automatically.
On the shared access, if you suspect there may be leftover triggers from a previous experiment, please contact us.
On a dedicated access, please remove any other triggers that may be active.
How can I find the best parameters for stimulation ? How should I stimulate to get maximal responses from the organoid?#
Our StimScan utility may be useful to find the best parameters for your experiment.
How long should I wait after a stimulation to start taking events into account and avoid artifacts?#
Excluding 10ms after the stimulation is a good rule of thumb, but it may vary depending on the stimulation parameters.
Please check the raw events’ shape and amplitude in the database to see if there are any artifacts, and adjust the delay accordingly.
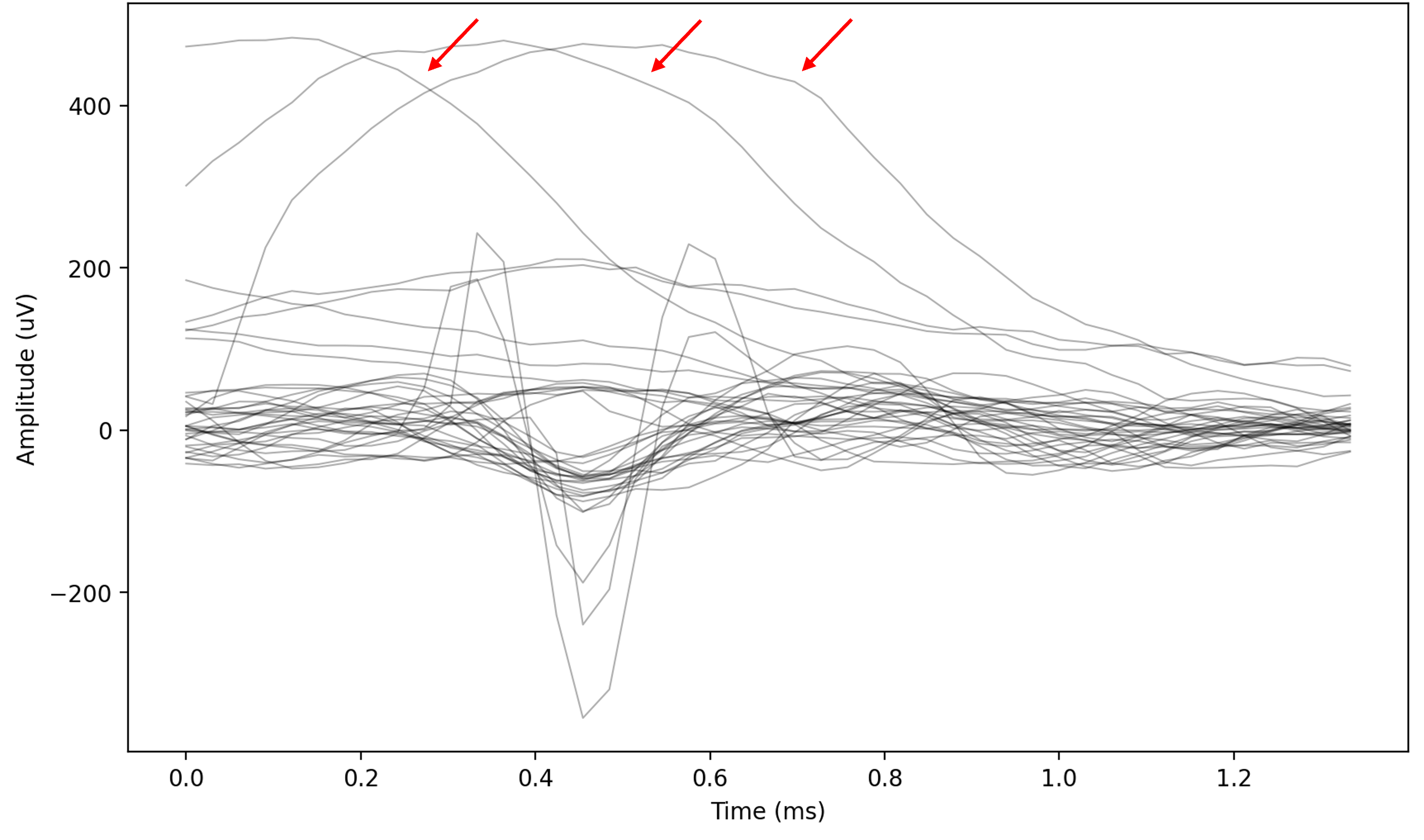
Fig. 11 Example of artifacts in the raw events. Notice the large “wave”-like shape, with a very large amplitude.#
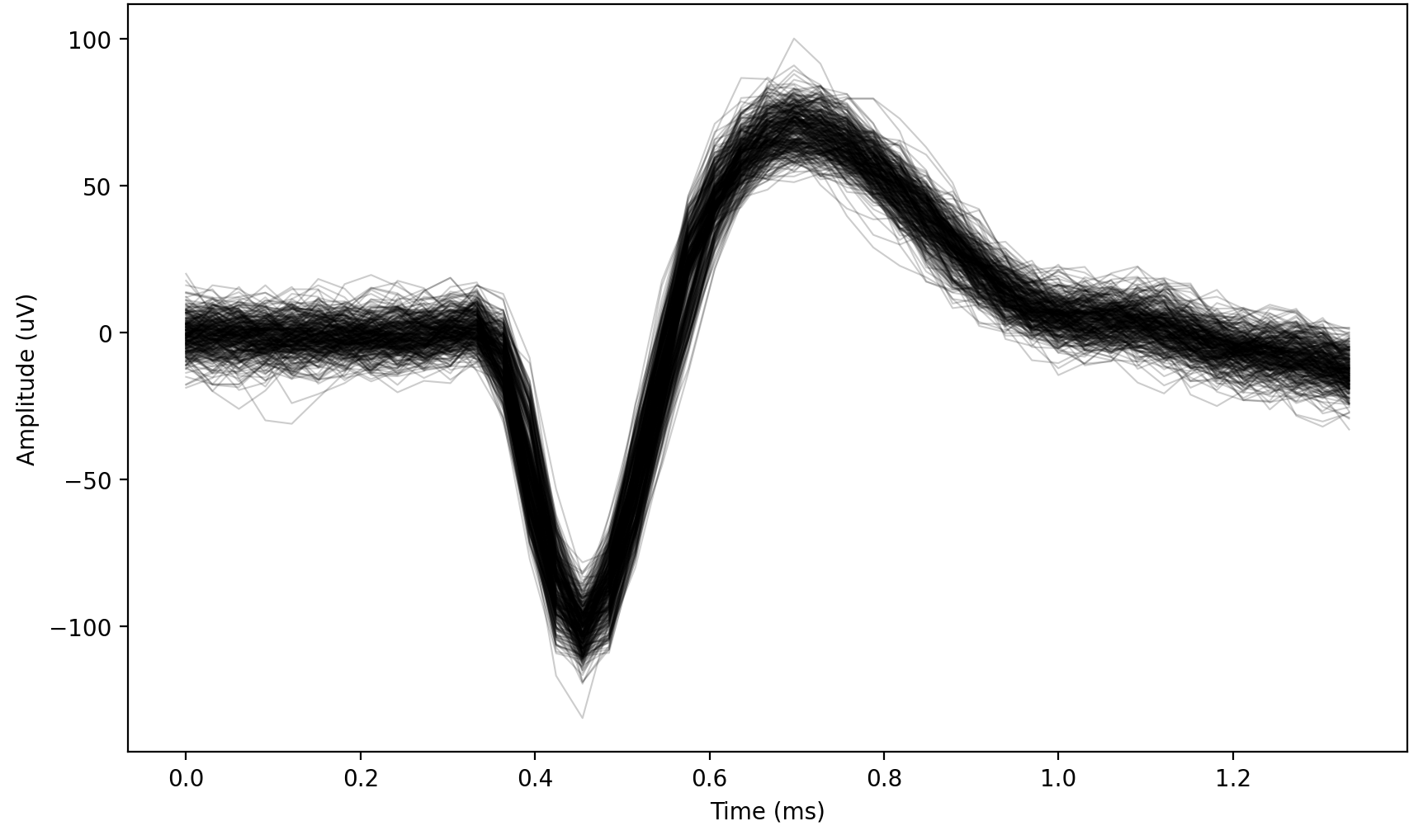
Fig. 12 Example of a signal from an organoid. Notice the variability, small amplitude, and “dip” before the peak.#
On a dedicated access, you can also tune analog and digital filters to decrease the delay; please contact us for more information.
I am unsure about the stimulation parameters and/or am not sure they are correctly set#
Our parameter visualization tool may help you check the parameters you are using.
If you are still unsure, please contact us.
Is it possible to perform high-speed closed loops, e.g. modulating stimulation frequency or changing parameters based on the activity of the organoid in real-time?#
Due to the network-based nature of the Neuroplatform, the delay between recording data directly on the Intan device and receiving it in your Python script will be at the strict minimum 35 to 50 ms.
Changing parameters currently takes 10 seconds on the shared access. On a dedicated access, parameters are changed as fast as the hardware allows.
We currently do not have the possibility to upload custom code to the Intan device, as such high-speed closed loops will be limited to the buffer rate of the software first, then the network delay, before it is available for processing in your Python script, then followed by network delays to send the stimulation back to the Intan device.
The network-based communication, the delay when changing parameters, and the use of Python make the current iteration of the Neuroplatform challenging for high-speed closed loops (< 200ms); however we are actively working on improving this aspect, as we are aware it is crucial to many experiments such as reservoir computing.
Stimulation does not elicit any response from the organoid#
Stimulations usually only elicit most of the response on their own channel, so make sure to check the correct channel.
As the organoid gets older, increasing the stimulation amplitude may be necessary.
Neurons require resources to fire, so fatigue may eventually occur after repeated stimulations. Use a higher delay between stimulations (1-10s) to avoid this issue.
If all else is ineffective, please contact us so we can change the organoids.
Database and experiment tokens#
How are spike events obtained ? Why is the minimal inter-spike interval always 3ms ?#
Events are recorded whenever the voltage crosses the event threshold of \(6\sigma\). This means that whenever the voltage crosses the threshold, an event is recorded for 3ms. As such, several spikes may be merged under several events, especially during bursts.
Caution
By default, the threshold is dynamic, which means it is computed based on the standard deviation of the noise at a certain time interval.
During bursts or stimulation, this standard deviation may increase, which will overall lower the amount of events recorded.
See Variable threshold for more information.
I do not have an Experiment token, or it is no longer valid#
Please contact us.
I receive only empty data/plots from the database, even though the same code worked previously#
On the shared access, there may have been a change of organoids, which requires you to change your experiment token. Please contact us so we can provide you with the updated token.
Neuroplatform utilities#
The LiveMEA recording tool stopped working#
We may have changed the way we stream data to the website. Please contact us.
There is an issue with Neuroplatform utilities#
Make sure you use Python 3.11 and have installed all the requirements for the specific utility you are using.
If you are still having issues or need help with upgrading your Python version, please contact us.


