Best Practices#
Here is a list of best practices for using the NeuroPlatform.#
These guidelines are designed to help you make the most of your experimental data, ensure consistent and reliable results, and build a solid foundation for your research.
We strongly encourage all users—especially newcomers—to follow these recommendations from the very beginning of their work on the platform. Adopting these habits early on will greatly improve your understanding of the data, reduce avoidable errors, and support reproducibility across experiments!
Download best practices cheatsheet : pdf link
Use of the Neuroplatform (Technical Integrity)#
🧪Checkpoint |
🔍 Why It Matters |
✅ Best Practices |
📊Data to Collect |
|---|---|---|---|
Proper login and logout procedures |
To prevent session conflicts |
Always follow the documentation guidelines to start and stop an experiment |
None (Make just sure to use the try / finally conditions as in the documentation for safety) |
Correct use of stim_params and triggers |
Hard to confirm post-experiment |
Review parameters before starting (You can use the param loader for that) |
Triggers, electrodes, if the enable condition is True (+ all parameters like amplitude, duration…) |
Have you verified your stimulation parameters? |
Hard to confirm post-experiment |
Use the param loader, keep track of your parameters |
Electrodes you stimulated, with which parameters (amplitude, duration, …) |
Are you sure that your stimulation parameters are optimal? |
Good stimulations are necessary to get good signals |
Apply a Grid Search of parameters (try different amplitudes, durations, electrodes…) and evaluate the responses to choose the best ones before to start |
Responsiveness for different parameters combinations |
Understanding the Model and Biological Context#
🧪Checkpoint |
🔍 Why It Matters |
✅ Best Practices |
📊Data to Collect |
|---|---|---|---|
Do you know the age of the organoid on the MEA? |
Activity varies significantly with age |
Check organoid age (you can do so by getting all events data for a specific organoid and checking the date of the first recording) and compare the results you got at different moments |
Age of the organoid on the MEA at each experiment |
Have you considered the initial bursting phase on MEA? |
Activity is often unstable during the first hours |
Wait at least a few hours before initial recording, check visually the organoid activity to assess burst |
Bursts presence (easy to see on the live on the FinalSpark website) |
Is the natural decline in activity accounted for? |
Activity may decrease independently of experiments |
Compare with inactive controls or track activity over time |
Comparison of results at different times |
Do you have the organoid token (FSxxx) ? |
Results may vary a lot between two batches due to biological variability, so better to know which organoid was concerned by which experiment |
Keep the token of each organoid you study |
Token of the organoid (FSxxx) Using this token, we can then find out which organoid/ medium/ set-up was used and improve data explainability in some cases (why does this one work better than another?) |
Data Validity#
🧪Checkpoint |
🔍 Why It Matters |
✅ Best Practices |
📊Data to Collect |
|---|---|---|---|
Have you reviewed raw data and spike shapes? |
Artifacts and real spikes look different |
Visualize raw data to discriminate spikes. Make an overlay of spikes! (The signals from one neuron should always have almost the same shape! If the overlay is bad, it may be noise) |
Overlay of spikes shape - Make sure to have a defined pattern, and not completely random signals. Make sure to have ‘biologically consistent’ data (a spike is usually within 100 uV, 1 ms) – see example below |
Do you compare spontaneous and evoked activity? |
Determines the effectiveness of stimulation, avoid to count spontaneous activity |
Compare data before and after a stimulation |
Metric that takes in account the spontaneous activity of the organoid when computing the elicited activity (for example, subtract the number of spikes happening within the 200 ms before the stimulation to the 200ms post-stim responses ) |
Have you checked for stimulation artifacts? |
Early peaks may be electrical artifacts |
Be cautious of latencies under ~10 ms after a stimulation |
Clean data without stimulation artifact (remove every spikes within 10ms post-stimulation or assess their raw shape) |
Are identical spikes appearing on all electrodes? |
May indicate a system artifact |
Analyze spatial and temporal propagation |
All the electrodes activity and not only the ones you are working with. Study correlations. |
Have you assessed electrode quality beforehand? |
Some electrodes may be noisy or inactive |
Exclude channels lacking clear signals prior to analysis |
State of each electrodes |
Is the experiment replicated on the same organoid? |
Tests for temporal variability (activity tends to decrease with time) |
Repeat experiments with controlled intervals |
Comparison of several trials of each experiment |
What is the interval between sessions? |
Activity may be affected by fatigue or plasticity |
Allow breaks between intense stimulations |
Responsiveness of the organoid post-stimulation - add breaks accordingly (usually at least a few seconds after each electrical stimulation patterns IF your goal is to come back to a ‘resting state’) |
Is the experiment replicated on different organoids? |
Necessary for generalizing effects, take in account biological variability |
Include multiple organoids |
Comparison of results though different organoids |
Have you implemented negative controls? |
Confirms the observed effects |
Include at least one control or baseline in each protocol |
Baseline of activity of the organoid before to start the experiment |
Examples of overlays :#
Here is a good spike signal that is consistent and seems to be produced by the same neuron.
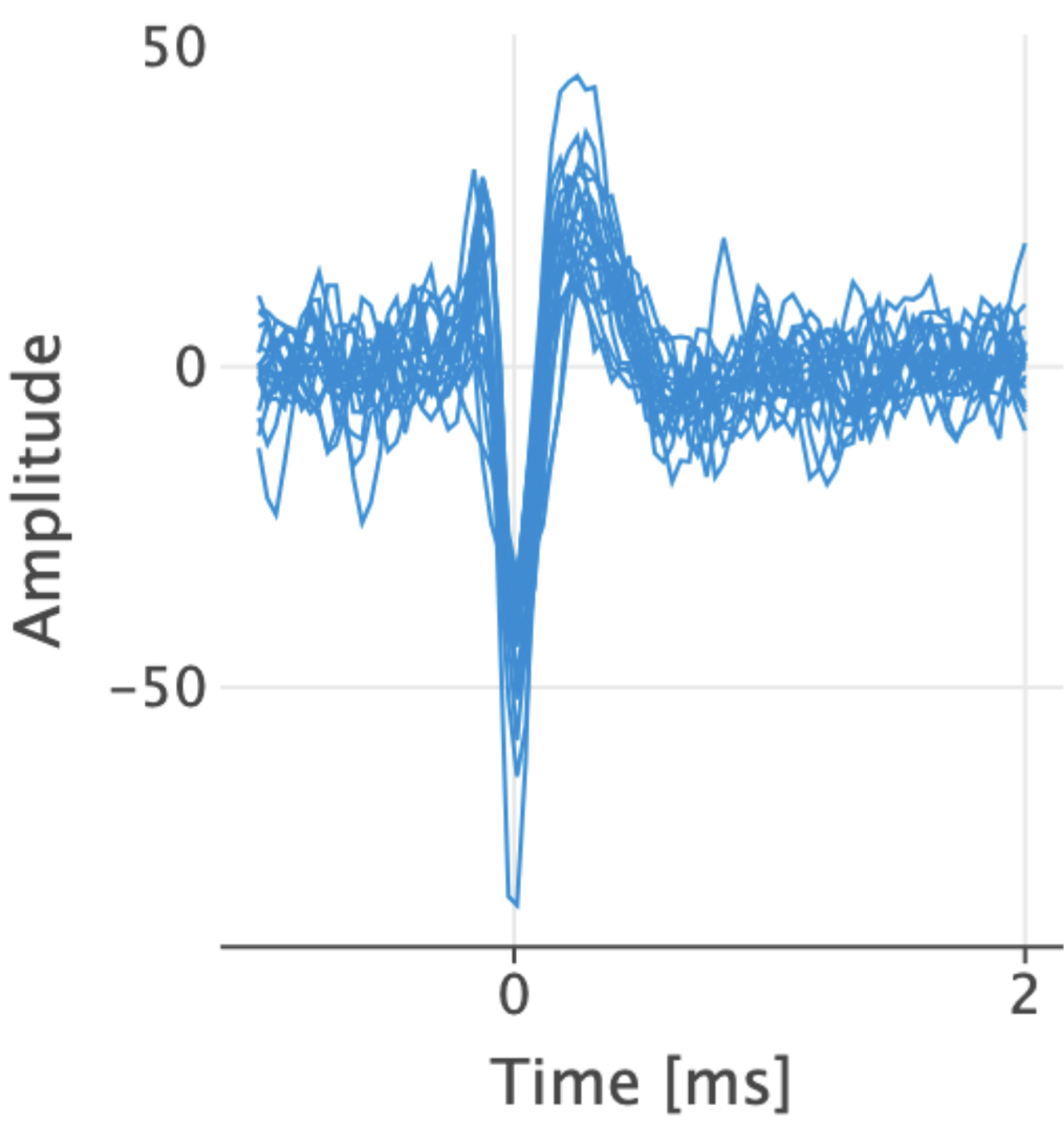
Fig. 5 Overlay Example 1#
Here is a good spike signal too but at least two neurons are involved to produce two spikes in less than 2ms.
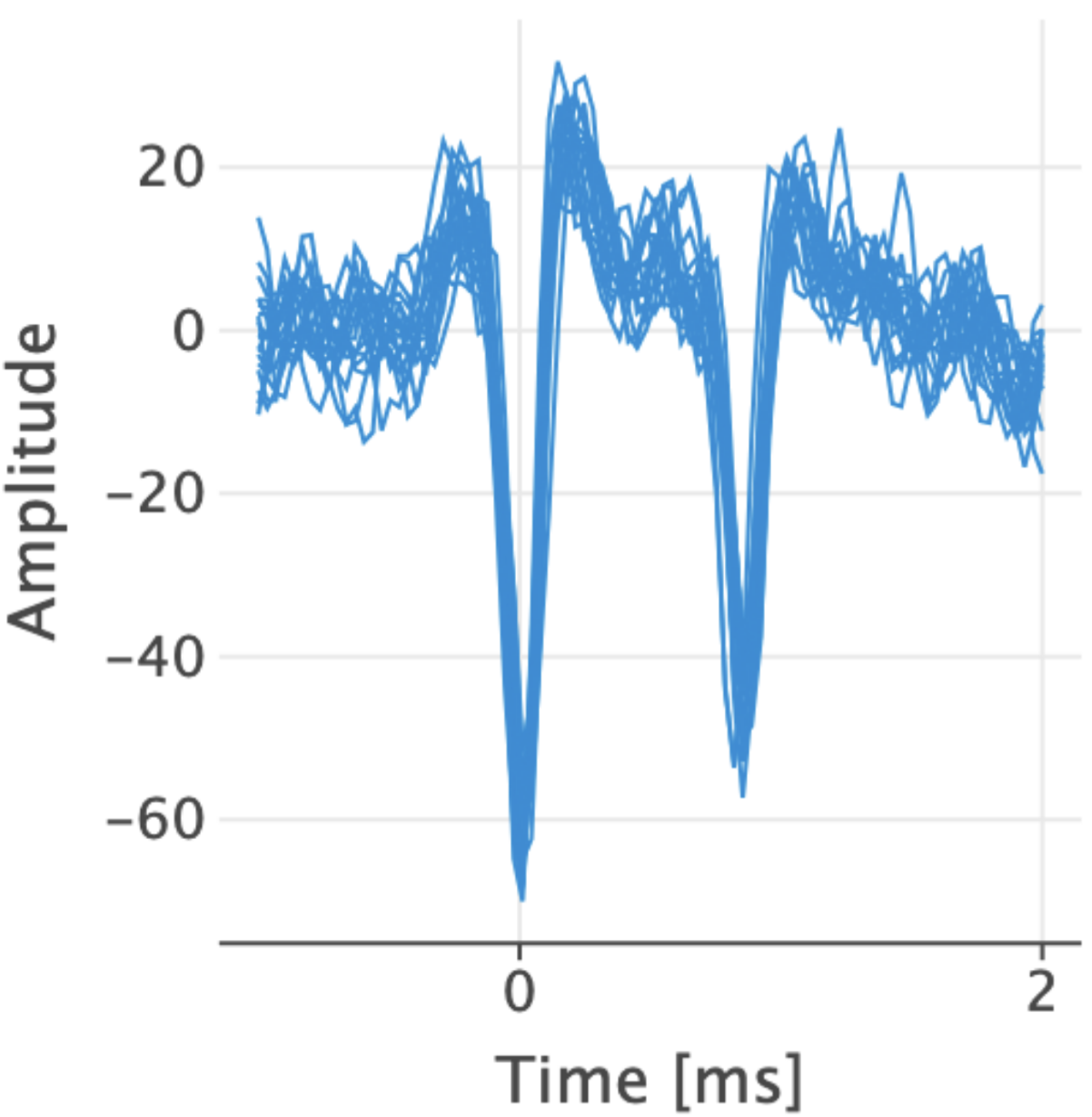
Fig. 6 Overlay Example 2#
